Research Overview
The Genetics of Tissue Regeneration
Most human tissues damaged by traumatic injury or disease do not regenerate, and current strategies to promote regeneration are limited. The ability to extensively repair and regrow damaged or missing tissue occurs frequently in other species, even in equivalent organs that have little to no regenerative capacity in people. Understanding what gives a tissue the ability to regenerate is an essential step towards developing therapeutic interventions to improve healing outcomes. However, the basic underlying genetic events that permit damage-induced regeneration remain unknown. This includes both the identities of factors required for tissue regrowth, and the regulatory events that lead to their appropriate activity throughout the regeneration process. Thus, the overall goal of our lab is to characterize the fundamental genetic mechanisms that allow tissues to regenerate, with a view to developing methods to enhance regenerative ability.
In many organisms, tissues frequently lose regenerative capacity as they mature from juvenile to adult stages. This phenomenon provides an ideal opportunity to investigate regeneration, as a single tissue can be examined at times of high and low regenerative capacity, and the differences responsible for successful regrowth can be identified. However, many of the organisms that lose regenerative capacity in this way are unsuitable for laboratory study.

Therefore, in order to study regeneration, we are using the well established and exceptionally powerful genetic model organism Drosophila melanogaster, the common fruit fly. The larval organs of Drosophila, known as imaginal discs, are able to regenerate following various types of damage. However, this ability is limited to early larval life and is progressively lost as the larvae mature.
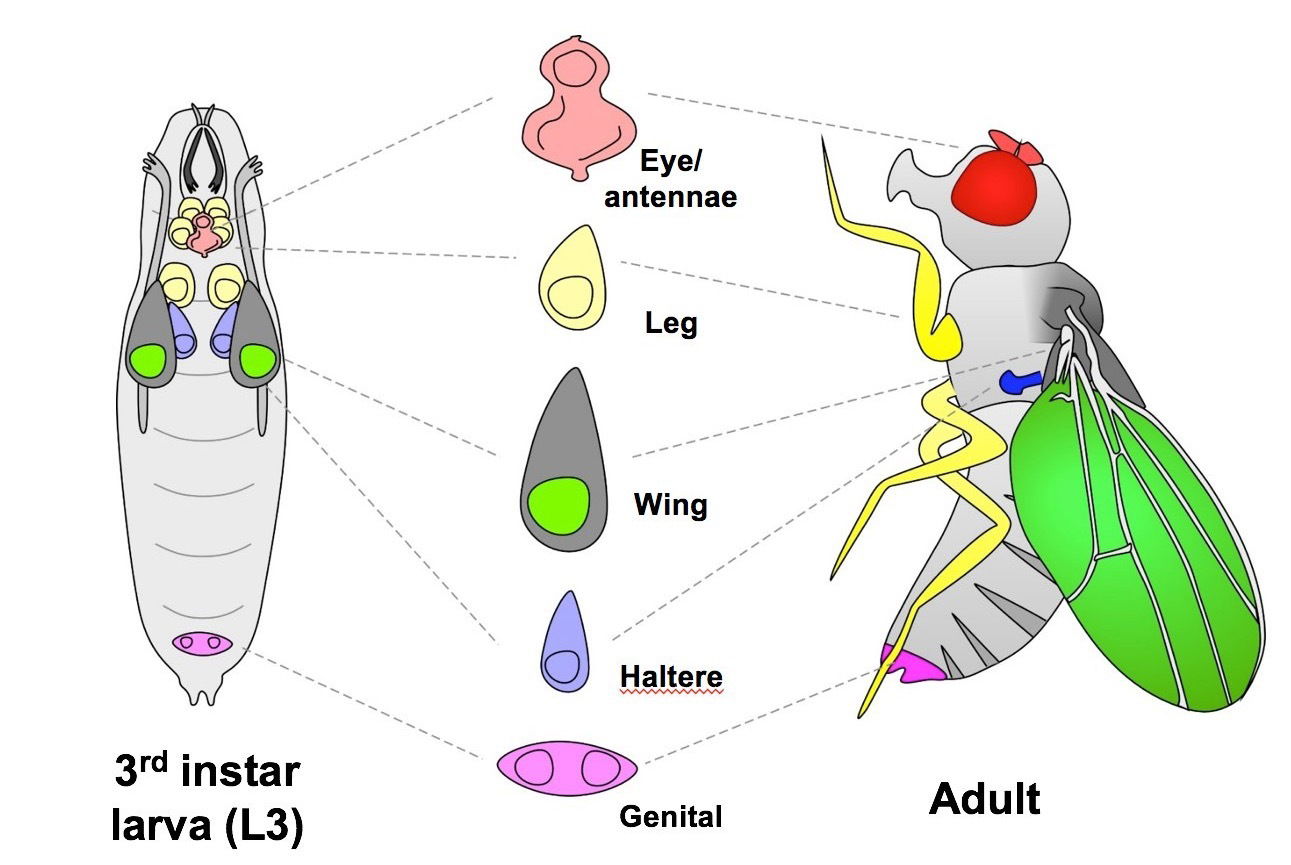
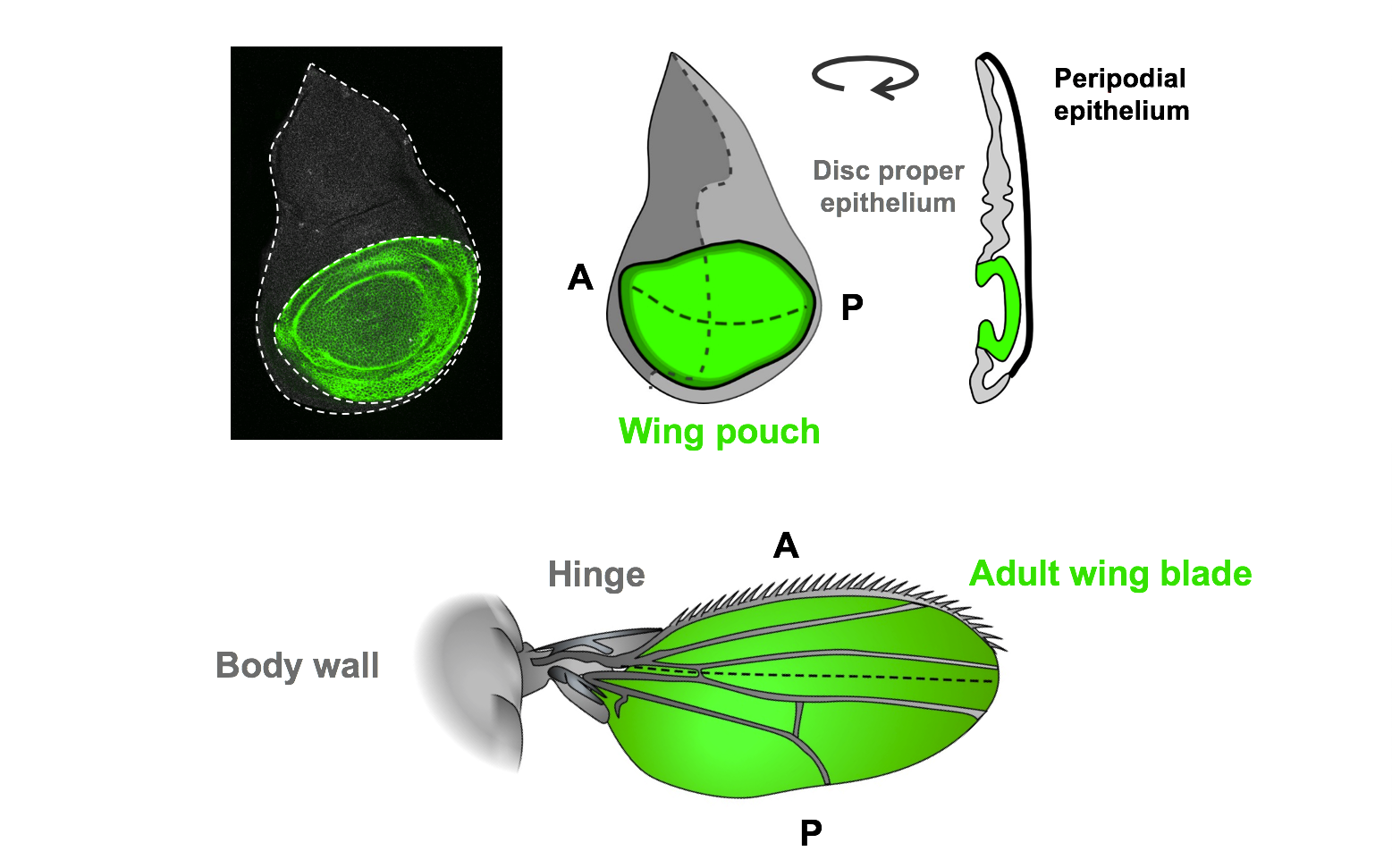
The wing imaginal disc, which forms the adult wing structures, is a non-essential organ that has been extensively studied as a model for tissue growth and development. The strong conservation of genetic pathways and the similarities in regenerative processes between Drosophila and mammals will ensure these findings will have relevance to human tissue regeneration.
Our previous research revealed the existence of a mechanism that controls the expression of genes involved in regeneration. We characterized a damage-activated region of the genome that, when activated by injury, leads to the expression of two nearby genes, WNT1 (wg) and WNT6. This Damage-Responsive and Maturity-Silenced (DRMS) enhancer induces expression of these genes only upon damage in younger tissues. As the tissue matures, the enhancer region becomes inactivated by epigenetic silencing, thus limiting damage-induced gene expression and contributing to the loss of regenerative capacity of this tissue. Importantly, this mechanism still permits developmental enhancers to shape expression of both genes in undamaged tissues, demonstrating how the regulation of a regeneration program can be separated from developmental gene expression.
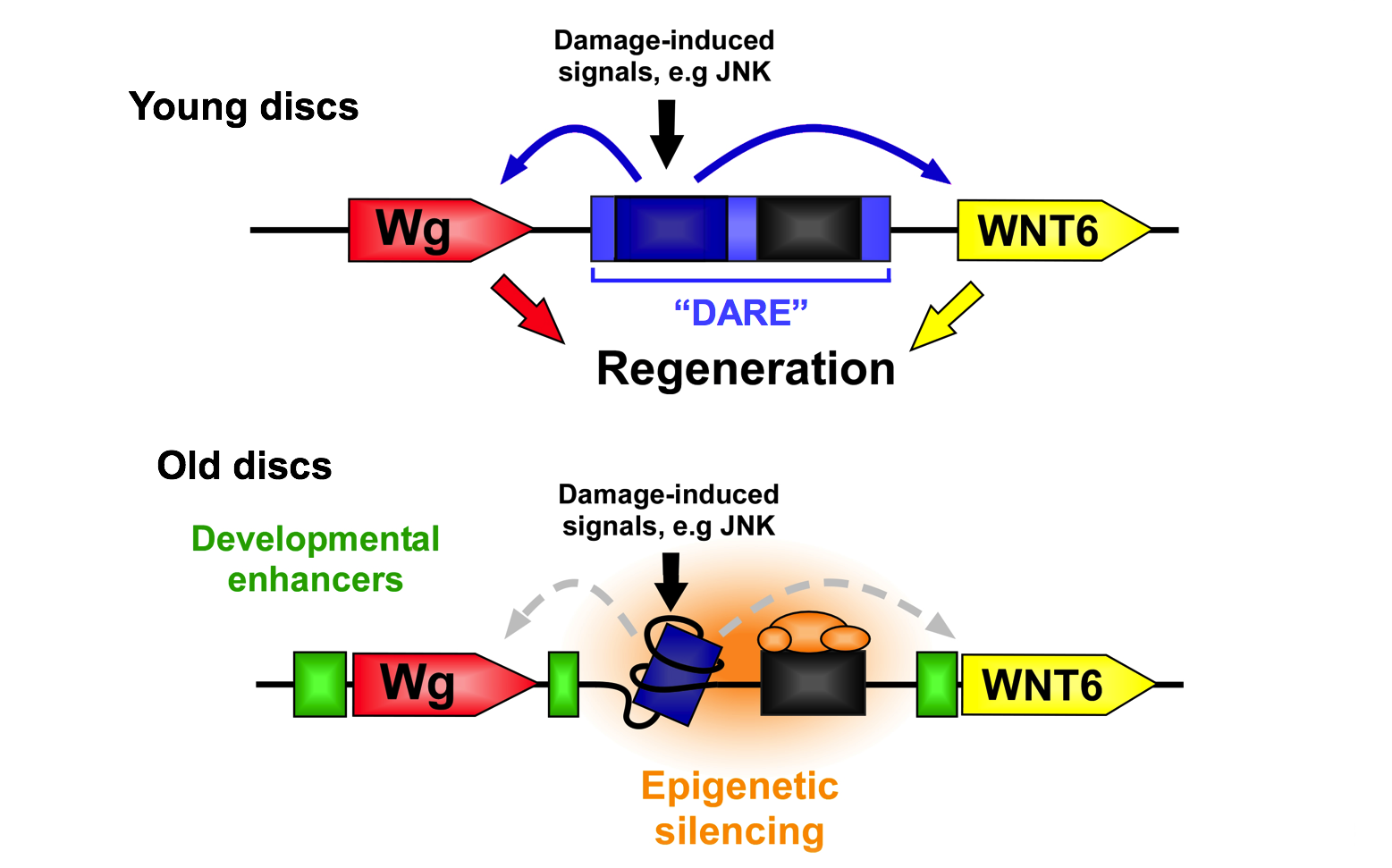
We have found that this mechanism is potentially widespread, controlling the expression of many other genes required for regeneration. Our work aims to characterize this mechanism, identify the target genes that might be regulated by it and are therefore involved in the regeneration program, and ultimately to elucidate genetic manipulations that can be used to augment regenerative ability. We are also interested in understanding how tissues respond to different types of cell death, specifically contrasting the regenerative response to necrosis versus apoptosis.